CEDA and JASMIN are facing two interrelated challenges regarding both Archive and user data: the data itself is growing rapidly year on year, and the introduction of new storage technology may require a change in workflows for users. Several software packages already exist to exploit this new storage technology, some of which involve re-writing the data into a new file format, which we believe is not suitable for archiving data. This article will present a data format, developed in conjunction with CEDA and NCAS CMS, that we believe can exploit the new storage technology and remain suitable as an archival data format.
Index
Background
The amount of data that we hold at CEDA and JASMIN, in both the CEDA Archive and the JASMIN Group Workspaces, is increasing rapidly year on year, in terms of the volume of data and the number of files. This is a problem faced by all data centres and, in response, the storage technologies used are expanding from a small set, such as disk and tape, to encompass newer technologies like Object Storage as well.
These newer storage technologies have different properties than the network attached, always available disk that is usually encountered on multi-user computing systems like JASMIN. This requires a different workflow and even a different way of thinking about data.
The storing of data has an inherent energy requirement and, unless 100% renewable energy is used, a corresponding Carbon Dioxide footprint. UKRI recently announced a commitment to zero carbon research by 2040. To reach this target, the CO2 footprint of data storage has to be reduced to zero. Even if renewable energy is abundant by then, it is still prudent to reduce the energy consumption of data storage. We believe that this can be achieved in (at least) two ways:
-
Reducing the storage required to hold the data, while still retaining the information content of the data. This involves optimising the data types used and also intelligent lossy compression. A future blog will write about this, but interested readers can look at these papers by Milan Kloewer and Charles Zender.
-
Reducing the duplication of data. At my previous research institute, I knew that portions of the CMIP5 dataset had been replicated on different servers in different departments around the campus. Sometimes it had even been replicated by different research groups in the same department. This duplication has both an energy use and e-waste implications, as the hard drives containing the data will need to be replaced over time. The reasons that the data is duplicated usually revolve around access:
- authorisation of access (who can access a particular server where the data is stored)
- search (finding the data required for an analysis)
- speed of access (how long does it take to complete an analysis)
Object Storage can help reduce the CO2 footprint of data by providing access via the internet to anyone who is authorised to read the data. This also makes it more suitable for use with Cloud Computing, as the data can be accessed from anywhere, not just from a machine that has mounted the disk. It provides the authorisation mechanism via a set of access keys and data can be read in parallel, mitigating some of the performance hit reading data across the network has, when compared to reading it from a disk.
As an illustration of this concept, Pangeo, ESGF and Google have made CMIP6 data available on the Google Cloud. This contains a repository of CMIP6 data, that has been converted to Zarr format, with an Intake catalogue that can be searched using Intake-esm. Scientists can use the popular Xarray data analysis software to read and analyse these datasets.
CEDA have also undertaken a similar project, using Xarray to convert CMIP6 data to Zarr and storing it on our on-premise Object Storage. The code repository for this project can be found here: CEDA CMIP6 Object Store, which also contains some examples of how to read the data from the Object Storage for JASMIN users.
A commercial system is available from the HDF-Group as the Highly Scalable Data Service.
Finally, as of version 4.8.0, the netCDF library supports reading and writing netCDF files in Zarr format.
It is apparent that in order to fully exploit the properties of Object Storage, new software is needed and that new file formats, or new ways of formatting the data while using an existing file format, may also be required. Zarr has its own custom file format that writes data out in “chunks”. As Zarr is a N-dimensional array based file format, these chunks are sub-domains of the array. Chunking data has the advantage that only the part of the dataset that is being analysed is read, as opposed to reading in the whole, potentially large, dataset. This is similar to how netCDF reads data from a disk and, as data from Object Storage is read across a network, it will be much faster to read than the equivalent un-chunked data. Chunking also allows the data to be streamed into memory, rather than caching to disk, as the smaller chunks can be held completely in memory, and also allow a parallel computing environment, such as Dask to be used.
By tailoring the file format and software to exploit the properties of Object Storage, Zarr can provide read and write performance approaching that of reading and writing netCDF files from and to disk, as can be seen in this paper by Xu, Paul and Banihirwe.
However, there are a few disadvantages to using Zarr that we believe make it unsuitable as a file format for the CEDA archive:
-
The data has to be transformed from the original file format (netCDF or something else) into Zarr, using Xarray or similar software. This requires the use of a super-computer or analysis cluster such as LOTUS on JASMIN, or the use of a cloud computing service, like Amazon AWS or Google Cloud. Either method incurs a cost in money, time, energy use and, potentially, CO2 emissions.
-
The beauty of netCDF as an archival file format is that it is self-describing. It contains details of the domain that a Variable is defined for; the dimensions of this domain; Variable, Dimension and global file metadata and the ability to form Groups for Variables and Dimensions and attach metadata to the Groups. There is no risk of this information becoming separated from the data it is describing as they are contained in the same file. Zarr stores metadata about the array it is encoding in a separate file (the
.zarrayfile), but it only stores information about the array, such as the shape, compression used, number of chunks and datatype. It does not contain the rich information a netCDF file does, like the Dimensions the array is defined over, or the metadata for the Variables and Dimensions. Xarray stores these in three other files:.zmetadata,.zattrand.zgroups. In a data archive, where data may be moved to tape, or migrated from one storage system to another, there is a real danger that these four files might become separated from the data they are describing. -
For a data archive, due to this risk of data and metadata becoming separated, it will be necessary to keep two copies of the data - the original for data preservation reasons, and the transformed data in Zarr format to allow performant analysis of the data. This is duplicating the data, which we want to avoid.
We believe that by retaining the file format of the original netCDF files, it is possible to create datasets, stored on Object Storage, that are both performant and suitable for long term archive. Instead we change the formatting of the data within the files and, possibly, creating new files. The basic concept is:
-
The chunked files,also known as Fragments, are netCDF files. They contain the Dimensions and domain for the data Variable in the Fragment, as well as the metadata for the Variable. The global metadata is replicated in each Fragment file as well.
-
A top-level control file contains the names and locations of the Fragments, as well as how the Fragments fit into the original data. This file is also in netCDF format and contains the Dimensions and domain for the whole data, and contains all of the metadata that the Fragment files contain.
S3netCDF demonstrates this concept to write and read data to and from both disk and Object Storage, via the Amazon S3 API. It uses an extended version (v0.5) of the Climate Forecast Aggregation conventions developed by our colleagues at NCAS CMS. Over the past year we have worked closely with our colleagures at NCAS CMS to standardise and consolidate the CFA conventions into a new version (v0.6). Work is now being undertaken to update S3netCDF to use these latest CFA convention.
The CFA Conventions
The CFA conventions formalise how a netCDF file can be used to describe a Dataset distributed across multiple other data files. It was originally designed to describe how a number of smaller files, for example a time series that is divided into decade long files, can be aggregated into a single Dataset. After consideration, it is apparent that this is the same problem encountered when splitting a large file into smaller Fragments (or chunks) to allow for good performance when reading and writing to Object Storage or cloud storage. At the highest level, it consists of two different types of files:
- Aggregation File - this is a netCDF file that contains the instructions needed to find a portion (sub-domain) of the data.
- Fragment File - this is also a netCDF file, which contains the data for the sub-domain.
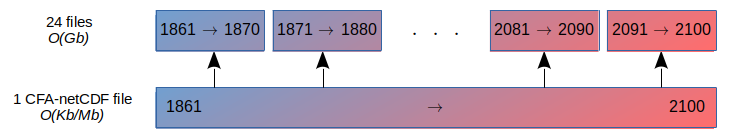
Figure 1 shows the relationship between these files for a timeseries of surface air temperature that has been split into 24 decade long files spanning the time period 1861 to 2100. Not only is it useful to view this as a 240 year long dataset, but splitting the data in this way provides an efficient way to distribute the Fragment Files to Object Storage (or cloud storage) so that the user only needs to access the time periods that they are interested in. The Aggregation File contains the instructions on where to find the Fragment Files, and where they fit into the timeseries.
The Aggregation File is a netCDF file that contains:
-
Aggregation Variable
A netCDF Variable that does not contain its own data, rather it contains instructions on how to create its data as an aggregation of data from other sources.
-
Aggregated Data
The data of an Aggregation Variable that exists as a set of instructions on how to populate an array from one or more other arrays stored elsewhere.
-
Aggregated Dimension
A netCDF Dimension that defines a Dimension of the Aggregated Data.
-
Fragment
An independent, possibly self-describing, array that defines a contiguous part of the Aggregated Data. The Aggregated Data is composed from a multi-dimensional orthogonal array of fragments.
-
Fragment Dimension
A Dimension of the multi-dimensional orthogonal array of Fragments that defines the Aggregated Data.
Figure 2 shows an example of one of these Aggregation Files. It contains one
Aggregation Variable temp that is defined on the time, latitude and
longitude Dimensions and has a domain size of 12 x 73 x 144. However, as
will be seen later, the definition of this is different than the definition of
Variables in a regular netCDF file.
In addition, Dimensions required by the CFA-Conventions are defined:
the Fragment Dimensions f_time, f_latitude and f_longitude and two extra
Dimensions. i defines the number of Aggregated Dimensions - here
there are three (time, latitude and longitude) and j defines the
maximum number of fragments per dimension. Here it is 2, as the time
Dimension is split into 2. and that is why the f_time Fragment Dimension has
length 2, whereas the f_latitude and f_longitude have length 1 - i.e.
there is no split for these Dimensions.
temp is the Aggregated Variable in this Aggregation File. It is defined as a
scaler variable - i.e. there are no Dimensions associated with it, like there
would be for a regular netCDF file. The Dimensions are, instead, specified in
the temp:aggregated_dimension Attribute, which defines the names of the
Aggregated Dimensions and their order. The Aggregated Variable also contains an
Attribute name aggregated_data, which contains a number of key: value pairs
and requires four keys to be defined: location, file, format and address.
The values are the names of the Variables that make up a part of the Aggregation
Instructions. In this example temp:aggregated_data = "file: aggregation_file"
indicates that, for the temp variable, the part of the Aggregation Instructions
that tell us which file a Fragment is stored in is contained in the netCDF
Variable aggregation_file.
Further down, these Aggregation Instruction Variables are defined. Their
Dimensions are the Fragment Dimensions (f_time, f_latitude, f_longitude)
and they each have a type (*string* or *int*). A complete set of Aggregation
Instructions requires four Variables:
-
aggregation_file : string. This provides the location of each Fragment. This may be a path to a file, or a URL to an OPeNDAP resource, or an AWS S3 Object. It may also reference the Aggregation File to indicate that the Variable is contained in the same file that the Aggregation Instructions are in.
-
aggregation_format : string. This indicates the format of the Fragment File. Currently only
ncis supported, which indicates a netCDF file. -
aggregation_location : int. This is an array indicating where the Fragment data fits into the Aggregation Variable data. In this example, the
timedimension has been split into two, whereas thelatitudeandlongitudehave not been split, and so have only one value. The_here indicates the missing value. The aggregation_location values are defined as contiguous spans of the Aggregation Variable data array, and the values give the length of each fragment. Therefore, the first fragment contains data for the0to5timeindices ([0:6]in Python) and the second contains data for the6 to 11indices ([6:12]in Python). -
aggregation_address : string. This defines the name of the netCDF Variable in the aggregation_file that contains the Fragment data. In this example we can see that the Variable name is
tosin both files.
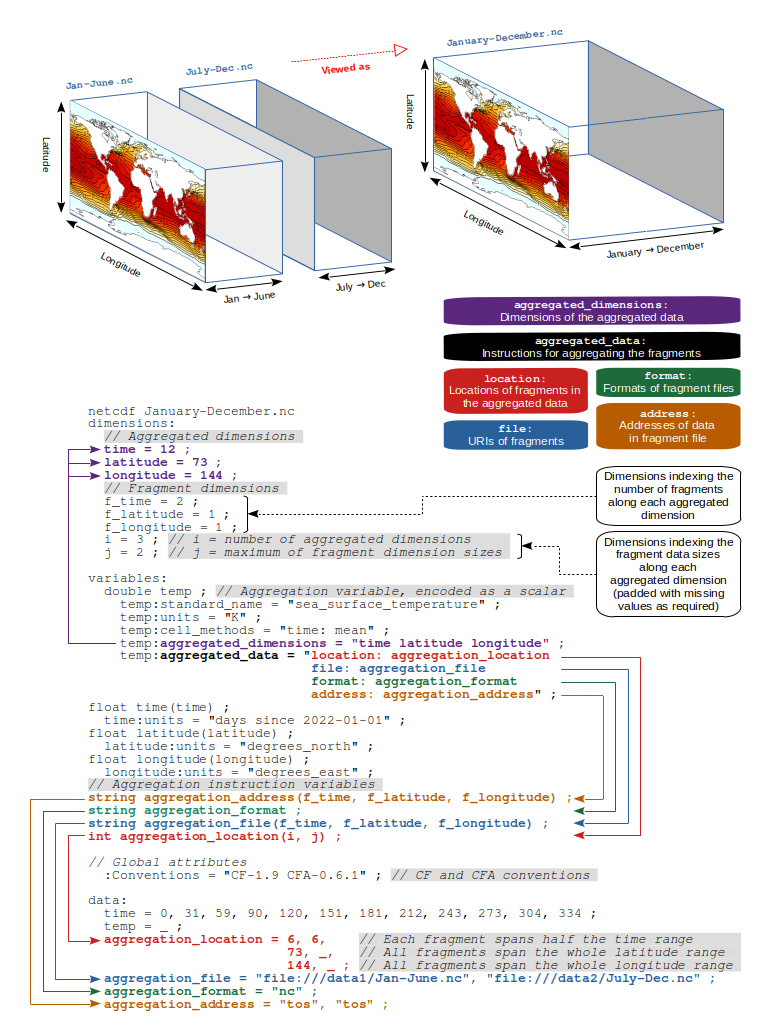
As can be seen in the example in Figure 2, the Aggregation File contains all
of the information necessary to record an aggregation of a number of files, or
the splitting of a larger file into a number of smaller files, and provide the
information on how to (re)construct the larger file from the smaller files. It
also contains all of the original metadata that is required for a
CF compliant netCDF file, including the units
for each Dimension, the standard name, units and any cell methods for the
Aggregation Variable, and any global metadata, such as the Conventions.
Although not stipulated by the CFA-Conventions, it is our intention that every
Fragment file is self-describing, contains the Dimension Variables for the
sub-domain that the Fragment is defined over, and contains the metadata for
the Aggregation Variable. In the example in Figure 2, the file:
file:///data1/Jan-June.nc will contain:
- a
latitudeDimension of size73and the correspondinglatitudeVariable with the latitude values in it. - a
longitudeDimension of size144and the correspondinglongitudeVariable with the longitude values in it. - a
timeDimension of size6and the correspondingtimeVariable with the values :[0, 31, 59, 90, 120, 151]. - the metadata for the
latitude,longitudeandtimeDimensions. - the metadata for the
latitude,longitudeandtimeVariables. - the global metadata.
It is this property of self-describing Fragments that we believe is crucial for a file format that is suitable for archival use. If any of the Fragments go missing, then we know exactly where that Fragment should fit into the domain of the Aggregation Variable. Similarly, if the Aggregation File (with all the Aggregation Instructions in it) is lost, then it can be reconstructed from the Fragments.
Conclusion
We have presented a data format that we believe has a number of advantages when it is used to reference data that is stored across different storage technology, such as Object Storage and disk. The CFA-Conventions allow the aggregation of existing files into a single Dataset, or the splitting of a large Dataset into smaller Fragment Files, while retaining the domain information and the metadata for the Variable, Dimensions and global metadata. The beginning of this article outlined how we think Zarr is not suitable as an archival data format, due to three disadvantages which we believe that using the CFA-Conventions overcomes:
-
Existing netCDF files do not have to be transformed into a new file format. An Aggregation File has to be generated, containing the Aggregation Instructions as to how the existing netCDF files fit together into a Dataset. For example, CMIP6 contains control runs of 500 years, split into 120 x 5 year long files. These can be aggregated into one Dataset, using the CFA-Conventions by generating the Aggregation Instructions to indicate that each 5 year long file belongs to an Aggregation Variable. It must be said, however, that the shape and size of the existing netCDF files may not be optimum for the best performance when analysing data and it may be necessary to refactor the Fragments into a new set of Fragments, which will have a similar processing burden as transforming the data to Zarr.
-
Each Fragment File contains the metadata from the original file, whether it is split or aggregated and also contains the domain information. There are no external files to be lost, deleted or corrupted.
-
Due to point 2, only one copy of the data needs to be kept, removing the need for data-duplication.
A future Tech Blog post will discuss how S3netCDF uses the CFA-Conventions to split large files into smaller Fragments, distributes the Fragments to disk, public cloud, OPeNDAP or Object Storage and then reads the Fragments in an efficient manner during an analysis, using minimal memory and exploiting the full network bandwidth.
AGU Presentations
A presentation on the CFA conventions was made at AGU 2021, as an interactive poster:
Acknowledgements
Many thanks to the team who worked on Version 0.6 of the CFA-Conventions:
- David Hassell - the leader and instigator of the CFA-Conventions
- Sadie Bartholomew
- Jonathan Gregory
- Bryan Lawrence
S3netCDF was supported through the ESiWACE and ESiWACE 2 projects. The projects ESiWACE and ESiWACE2 have received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreements No 675191 and 823988.

 Neil Massey
Neil Massey